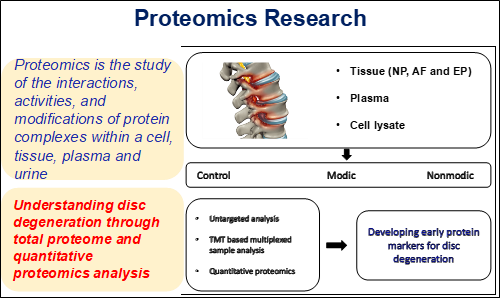
The GRC Proteomics Laboratory is established to understand the disc degeneration at molecular level using state-of-the-art mass spectrometry based platform which was supported by Department of Biotechnology, New Delhi. Researchers mainly work on the proteins from disc tissue and plasma of normal and degenerated patients. Main focuses of proteomics research include complete understanding of the pathophysiology of intervertebral disc diseases and development of biomarkers that would help in the prognostic approach of the disease.
Proteomics contribute to the analysis of complex proteins which reflects the dynamic responses inside the cells at a specific point of time encoded by a genome. This dynamic nature justifies studying gene expression of the disease state through proteins; in addition proteomics studies extend its analysis on post-translational modification, spatial interaction between cells and their signaling events. Hence, our research laboratory progresses towards achieving the long term goal of developing clinical biomarkers for early diagnosis of the disc disease. Our research work focuses on the project “To unravel the molecular events contrasting between biological aging and degeneration of IVD through proteomics”.
Protein profiling
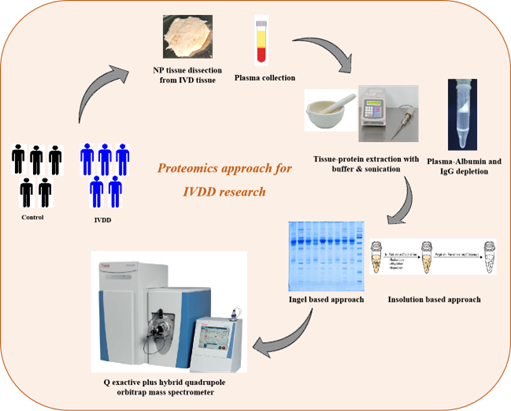
Research at proteomics involves analysis and identification of disc tissue and plasma proteins from intervertebral disc degenerative patients. Our work flow involves extraction of proteins from the disc tissues. Plasma proteomics involves depletion of top two abundant proteins including albumin and IgG which mask the identification of low abundant proteins using depletion kits. We employ in-gel or in-solution based tryptic digestion for mass spectrometric identification of disc tissue and plasma proteins. The figure below denotes the workflow followed for proteomics approach to understand IVDD.
Targeted analysis
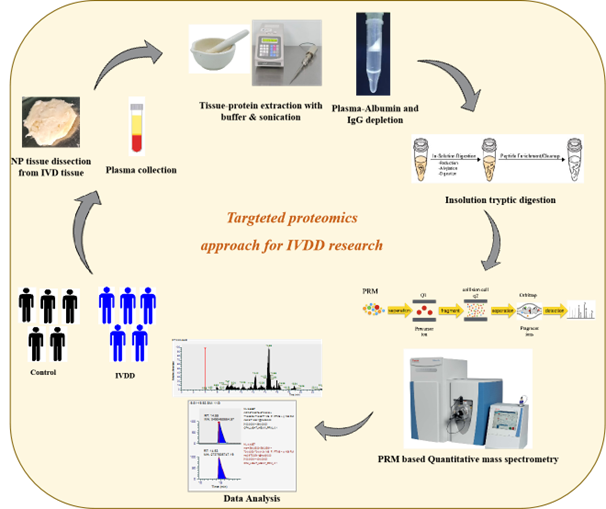
Targeted quantitation is required to verify and validate the selected candidate biomarkers for disease diagnosis or prognosis. Targeted proteomics via selected reaction monitoring (SRM) or parallel reaction monitoring (PRM) enables fast and sensitive detection of a preselected set of target peptides. Parallel-reaction monitoring (PRM) provides high selectivity, high sensitivity, and high-throughput quantification with confident targeted peptide confirmation. PRM is most suitable for quantifying tens to hundreds of targeted proteins in complex matrices. Mass spectrometer specifically capture target precursor ions, fragments the precursors, and then detects all resulting product ions in the Orbitrap mass analyzer. Quantification is carried out using selected fragment (product) ions post data acquisition. Advantages include more accuracy and attomole-level limits of detection and quantification. It also enables the confident confirmation of the peptide identity with spectral library matching. We employ Thermo Scientific Q Exactive™ hybrid quadrupole-Orbitrap instrument for PRM analyses to validate our discovery proteomics findings in tissue and plasma of IVDD.