It is the essential computational and statistical engine for managing, analyzing, and interpreting the massive and complex datasets generated by "omics" technologies. It transforms raw data from genomics, proteomics, and other fields into meaningful biological insights, which are then applied in areas such as disease diagnosis, personalized medicine, and drug development.
Research in Bioinformatics
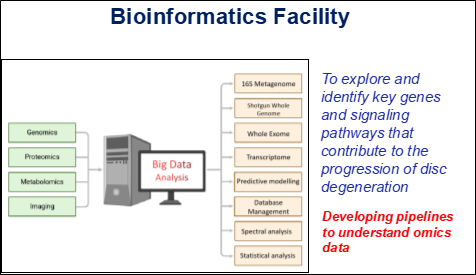
The facility unifies genomic, metagenomic, transcriptomic, proteomic, metabolomic, microbiome, and imaging datasets to obtain a holistic picture of disc health across the disease spectrum. By layering these diverse data types, the group moves beyond purely descriptive work toward mechanistic insight and predictive frameworks that can inform diagnosis, prognosis, and individualized therapeutic strategies for people with disc degeneration.
Core research activities aim to delineate molecular signatures that differentiate healthy from degenerated discs, emphasizing inflammatory cascades and host–microbiome crosstalk. Through coordinated analysis of genes, proteins, metabolites, and microbial communities, the team seeks early indicators of disease onset, progression, and treatment response. Metagenomic and metatranscriptomic investigations highlight perturbed pathways, regulatory circuits, and microbiome shifts reveal downstream functional consequences within disc tissues and their surrounding milieu. These molecular readouts are interpreted alongside imaging findings and clinical data to clarify how subtle biological changes culminate in structural damage and symptomatic disease.
Current initiatives focus on robust analytical frameworks for 16S microbiome surveys, whole-genome and exome sequencing, metagenomic datasets, and transcriptome profiling from disc-related biospecimens. Tailored workflows encompass stringent quality control, feature extraction, quantification, and multi-layer functional interpretation, ensuring consistent, transparent handling of complex datasets.The group also develops prognostic and predictive models that integrate multi-omics features with imaging metrics and clinical variables to classify patients and anticipate disease trajectories. Sophisticated statistical methods and machine-learning strategies are used to uncover patterns that can support risk stratification, guide therapeutic choices, and monitor response over time.
Facilities and capabilities
The facility maintains a dedicated bioinformatics environment optimized for large-scale omics in disc degeneration research. This setup supports intensive data processing, secure management of sensitive clinical information, and efficient implementation of state-of-the-art analytical workflows. Strengths span microbiome and metagenomic interpretation, protein-centric and network-evel analyses, integration of heterogeneous data types, and rigorous statistical modeling. Working closely with basic scientists, clinicians, and spine surgeons, the facility provides a strong platform for discovery science, translational studies, and emerging precision-medicine efforts in spinal health.